Choosing A Drug Design Strategy
Which strategy to choose when planning a drug discovery project? Maybe structure-based design, ligand-based design, or de novo design? These questions are discussed in our series on structure-based drug design in this second post.
This is the second post in our series on structural biology and structure-based drug design. Before the start, I would like to thank everyone who supported the project on LinkedIn through your “likes,” by starting to follow our company, and by sharing the link with your network. Thank you very much! This was encouraging! I generally think that we need more science on LinkedIn. It also occurred to me that writing short popular science articles could be a good way for Ph.D. students to tell the world some exciting stories and market themselves as potential future employees. Together, we could make science much more visible on LinkedIn!
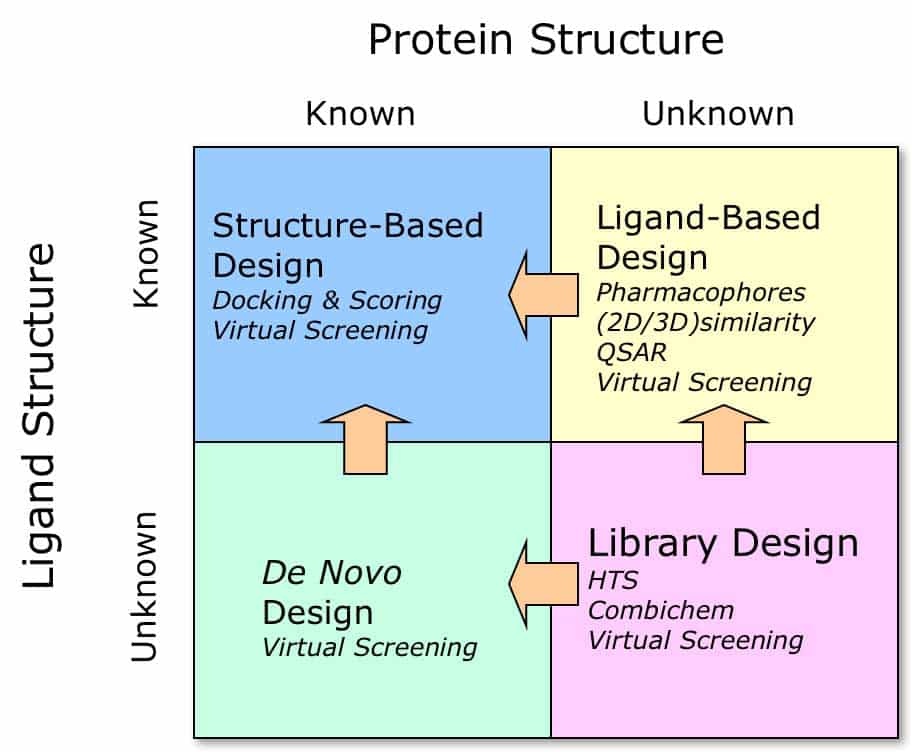
The previous post discussed the “when” question – when we need structural biology during drug discovery. Here, we will discuss another question, which often needs to be answered before a drug discovery project starts: a “what“ question. Assuming we have a target, what strategy must we follow to start a structure-based drug design project? You can see what type of information I mean on the image – it is the structure. The two axes on the image represent a combination of the two main parameters, known/unknown protein structure and known/unknown ligand structure. The general definition of “a ligand” refers to a molecule, most commonly a small molecule, that can modulate the target protein’s function after binding at a particular site. The binding site could be an active site (substrate binding site) if the target is an enzyme, or it could be a co-factor binding site, an allosteric site, or a protein-protein interaction site. Looking again at the image, we see four colored quadrants; inside each, we can read about the possible actions to follow:
Blue: known protein and ligand structures – here, we have all the advantages of structure-based drug design (SBDD); we can also do virtual screening, docking and scoring, and much more!
Green: known protein structure, unknown ligand structure – de novo design will be the starting point, and after identifying the first good quality binders, we can proceed with SBDD to get the method working to our advantage.
.
Yellow: known ligand structure, unknown protein structure – the so-called ligand-based drug design. Here, we can run screening using, for example, a targeted compound library, build a QSAR model (quantitative structure-activity relationships, SAR like in SARomics!), and create a pharmacophore model. If we can find a reliable predicted model of our target, we could even run a virtual screening. Hopefully, we will have sufficient data to start a structure-based drug design project at some stage.
Pink: Protein and ligand structures are unknown, which is probably the most time-consuming (and costly) situation. In my opinion, determining the 3-dimensional structure of the protein target would be much more efficient before proceeding with the project.
It is obvious where we want to be at the start of a project — it is in the blue quadrant! Structure-based drug design allows us to leverage all the power of a 3D structure, thereby accelerating the project and saving substantial costs.
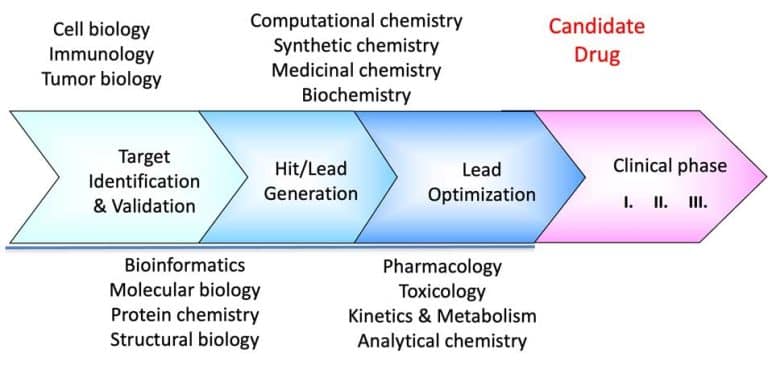
Our methods and technology center discusses structure-based drug discovery strategies in more detail. In a later post, we will discuss the following steps: fragment/compound screening, hit identification, hit expansion, and lead generation. As the image above shows, screening is central to any discovery strategy.
If you haven’t done that, follow us on LinkedIn, and stay tuned!