Publications co-authored by SARomics Biostructures’ team
At SARomics Biostructures, we showcase our recent publications below. You can follow the links or scroll down to see earlier publications. If available, the PDB code indicates our contribution to the work.
Please note that this list represents only a small fraction of all the projects handled by the company over the years. While some publications are the result of EU-funded projects where SARomics Biostructures participated as an industrial partner, most of them contain data from our customers. We are grateful to our customers for allowing our team members to be co-authors. However, the ownership and decision to publish the data lie entirely with the customers.
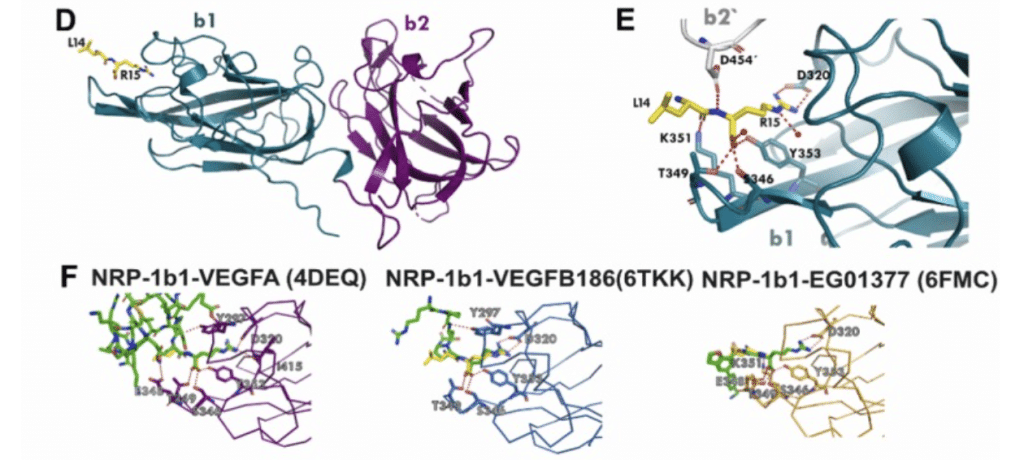
Chen Y, Gialeli C, Shen J, Dunér P, Walse B, Duelli A, Caing-Carlsson R, Blom AM , Zibert JR, Hultgårdh Nilsson A, Alenfall J, Liang C, Nilsson J (2024).
Identification of an osteopontin-derived peptide that binds neuropilin-1 and activates vascular repair responses and angiogenesis.
Pharmacological Res., V. 205, 107259
PDB entry:
9EOU – Crystal Structure of the b1b2 domains from Human Neuropilin-1 in complex with a peptide.
The osteopontin-derived peptide FOL-005 stimulates hair growth. Using ligand-receptor glyco-capture technology we identified neuropilin-1 (NRP-1), a known co-receptor for vascular endothelial growth factor (VEGF) receptors, as the most probable receptor for FOL-005 and the more stable analogue FOL-026. X-ray crystallography and microscale thermophoresis analysis revealed that FOL-026 shares binding site with VEGF in the NRP-1 b1-subdomain.
van Klaveren S, Hassan M, Håkansson M, Johnsson RE, Larsson J, Jakopin Ž, Anderluh M, Leffler H, Tomašič T, Nilsson UJ (2024).
Galectin-8N-Selective 4-Halophenylphthalazinone-Galactals Double π-Stack in a Unique Pocket.
ACS Med Chem Lett 15, 1319-1324.
PDB entry:
9FXZ – Galectin-8 N-terminal carbohydrate recognition domain in complex with 4-(bromophenyl)phthalazinone D-galactal ligand.
Galectin-8 contains two different carbohydrate recognition domains (CRDs). Selective inhibitors for at least one CRD are desirable for galectin-8 biology studies and potentially for pharmacological purposes. Structure-guided design led to the discovery of potent and selective glycomimetic–heterocycle hybrid ligands, with a 4-(p-bromophenyl)phthalazinone derivative displaying a 34 μM Kd for galectin-8N (N-terminal CRD), no binding to galectin-8C (C-terminal CRD), -1, -3, -4N, -7, -9C, or -9N, and >40-fold selectivity over galectin-4C. Selectivity was achieved with the halogenated 4-phenylphthalazinone moiety occupying a galectin-8N-specific sub-pocket. A 1.30 Å resolution X-ray structure revealed the phthalazinone moiety stacking with Arg45 and the 4-bromophenyl moiety stacking both Arg59 and Tyr141 of galectin-8N.

Malinge P, Chauchet X, Bourguignon J, Bosson N, Calloud S, Bautzova T, Borlet M, Laursen M, Kelpsas V, Rose R, Gueneau F, Ravn U, Magistrelli G, Fischer N (2024).
Structural analysis of light chain-driven bispecific antibodies targeting CD47 and PD-L1.
mAbs 16, 2362432.
PDB entry:
8RP8 – Structure of K2 Fab in complex with human CD47 ECD
8RPB – Structure of S79 Fab in complex with IgV domain of human PD-L1
In contrast to natural antibodies that rely mainly on the heavy chain to establish contacts with their cognate antigen, we have developed a bispecific antibody format in which the light chain (LC) drives antigen binding and specificity. To better understand epitope-paratope interactions in this context, we determined the X-ray crystallographic structures of an antigen binding fragment (Fab) in complex with human CD47 and another Fab in complex with human PD-L1. These Fabs contain a κ-LC and a λ-LC, respectively, which are paired with an identical heavy chain (HC). The structural analysis of these complexes revealed the dominant contribution of the LCs to antigen binding, but also that the common HC provides some contacts in both CD47 and PD-L1 Fab complexes. The anti-CD47 Fab was affinity optimized by diversifying complementary-determining regions of the LC followed by phage display selections.

Modulating protein unfolding and refolding via the synergistic association of an anionic and a nonionic surfactant.
Hjalte J, Diehl C, Leung AE, Poon JF, Porcar L, Dalgliesh R, Sjögren H, Wahlgren M, Sanchez-Fernandez A (2024).
J Colloid Interface Sci. 672, 244-255.
In this work, the behavior of three model proteins (human growth hormone, bovine serum albumin, and β-lactoglobulin) was investigated in the presence of the anionic surfactant sodium dodecylsulfate, the nonionic surfactant β-dodecylmaltoside, and mixtures of both surfactants. The transitions occurring to the proteins were determined using intrinsic fluorescence spectroscopy and far-UV circular dichroism. Based on these results, we developed a detailed interaction model for human growth hormone. Using nuclear magnetic resonance and contrast-variation small-angle neutron scattering, we studied the amino acid environment and the conformational state of the protein.

Zetterberg FR, Peterson K, Nilsson UJ, Andréasson Dahlgren K, Diehl C, Holyer I, Håkansson M, Khabut A, Kahl-Knutson B, Leffler H, MacKinnon AC, Roper JA, Slack RJ, Zarrizi R, Pedersen A (2024).
Discovery of the Selective and Orally Available Galectin-1 Inhibitor GB1908 as a Potential Treatment for Lung Cancer.
J Med Chem. 2024 Jun 13;67(11):9374-9388.
PDB entry:
4Q26 – Crystal Structure of Galectin-1 in Complex with N-Acetyllactosamine
We have previously described a new series of selective and orally available galectin-1 inhibitors resulting in the thiazole-containing glycomimetic GB1490. Here, we show that the introduction of polar substituents to the thiazole ring results in galectin-1-specific compounds with low nM affinities. X-ray structural analysis of a new ligand-galectin-1 complex shows changes in the binding mode and ligand-protein hydrogen bond interactions compared to the GB1490-galectin-1 complex. These new high affinity ligands.

Prospective de novo drug design with deep interactome learning.
Atz K, Cotos L, Isert C, Håkansson M, Focht D, Hilleke M, Nippa DF, Iff M, Ledergerber J, Schiebroek CCG, Romeo V, Hiss JA, Merk D, Schneider P, Kuhn B, Grether U, Schneider G (2024)
Nat Commun. 15, 3408.
PDB entry:
8PBO – Deep interactome learning for generative drug design
De novo drug design aims to generate molecules from scratch that possess specific chemical and pharmacological properties. The group of Professor Gisbert Schneider at ETH Zurich, presents a computational approach utilizing interactome-based deep learning for ligand-based and structure-based de novo design of drug-like molecules in this publication. New ligands targeting the binding site of the human peroxisome proliferator-activated receptor subtype gamma (PPARgamma, a protein from our FastLane™ Premium library) were generated. The system considers synthesizability, novelty, bioactivity, and physicochemical properties for ligand design. The binding mode of the ligand was confirmed by the crystal structure of the protein-ligand complex provided by the SARomics team. The structure of the complex shows that the ligand effectively interacts with the receptor in a canonical binding mode, while also demonstrating the desired selectivity towards the receptor and favorable ADME properties.

Zetterberg FR, Diehl C, Håkansson M, Kahl-Knutson B, Leffler H, Nilsson UJ, Peterson K, Roper JA & Slack RJ (2023).
Discovery of Selective and Orally Available Galectin-1 Inhibitors.
J. Med. Chem. https://doi.org/10.1021/acs.jmedchem.3c01787
PDB entries:
8OJP – Human galectin 1 in complex with inhibitor
A new series of orally available α-d-galactopyranosides with high affinity and specificity toward galectin-1 have been discovered. High affinity and specificity were achieved by changing six-membered aryl-triazolyl substituents in a series of recently published galectin-3-selective α-d-thiogalactosides (e.g., GB1107 Kd galectin-1/3 3.7/0.037 μM) for five-membered heterocycles such as thiazoles. One compound, GB1490 (Kd galectin-1/3 0.4/2.7 μM), was selected for further characterization toward a panel of galectins showing a selectivity of 6- to 320-fold dependent on galectin. The X-ray structure of GB1490 bound to galectin-1 reveals the compound bound in a single conformation in the carbohydrate binding site.

Boonkrai C, Cotrone TS, Chaisuriyong W, Tantawichien T, Thisyakorn U, Fernandez S, Hunsawong T, Reed M, Wongtangprasert T, Audomsun T, Phakham T, Attakitbancha C, Saelao P, Focht D, Kimbung R, Welin M, Malik AA, Pisitkun T & Srisawat N (2023)
Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
PLOS ONE 18(5): e0284173, https://doi.org/10.1371/journal.pone.0284173
PDB entries:
8BSE– Crystal structure of SARS-COV-2 receptor binding domain (RBD) in complex with 1D1 Fab.
8BSF – Crystal structure of SARS-COV-2 receptor binding domain (RBD-beta variant) in complex with 3D2 Fab.
This study aimed to develop a monoclonal antibody against SARS-CoV-2 from B cells of recovered COVID-19 patients, which might have beneficial therapeutic purposes for COVID-19 patients. The authors generated human monoclonal antibodies (hmAbs) against the receptor binding domain (RBD) protein of SARS-CoV-2 using developed hybridoma technology. The isolated hmAbs against the RBD protein (wild-type) showed high binding activity and neutralized the interaction between the RBD and the cellular receptor angiotensin-converting enzyme 2 (ACE2) protein. Epitope binning and crystallography results displayed target epitopes of these antibodies in distinct regions beneficial in the mix as a cocktail.

Liu CY, Ahonen CL, Brown ME, Zhou L, Welin M, Krauland EM, Pejchal R, Widboom PF & Battles MB (2023).
Structure-based engineering of a novel CD3ε-targeting antibody for reduced polyreactivity
MAbs 15: 2189974-2189974. https://doi.org/10.1080/19420862.2023.2189974
PDB entries:
8F0L – Crystal Structure of the Human T cell Receptor CD3(EPSILON) N-Terminal Peptide Complexed with ADI-26906 FAB.
Bispecific antibodies continue to represent a growth area for antibody therapeutics, with roughly a third of molecules in clinical development being T-cell engagers that use an anti-CD3 binding arm. Using insights from the crystal structure of anti-Hu/Cy CD3 antibody ADI- 26906 in complex with CD3ε and antibody engineering using a yeast-based platform, the authors have derived high-affinity CD3 antibody variants with very low polyreactivity and significantly improved biophysical developability. Comparison of these variants with CD3 antibodies in the clinic (as part of bi- or multi- specifics) shows that affinity for CD3 is correlated with polyreactivity. The engineered CD3 antibodies break this correlation, forming a broad affinity range with no to low polyreactivity. Such antibodies will enable bispecifics with improved pharmacokinetic and safety profiles and suggest engineering solutions that will benefit the large and growing sector of T-cell engagers.

Simon IA, Bjørn-Yoshimoto WE, Harpsøe K, Iliadis S, Svensson B, Jensen AA, Gloriam DE (2023).
Ligand selectivity hotspots in serotonin GPCRs.
Trenda Pharm. Sci. 44, 978-990. https://doi.org/10.1016/j.tips.2023.09.012
Serotonin is a neurotransmitter involved in the regulation of numerous physiological processes. Its action is also modulated by drugs in the treatment of schizophrenia, depression, migraine, and obesity. However, these drugs typically have adverse effects caused by promiscuous binding across 12 serotonin and more than 20 homologous receptors. This review maps 19 ‘selectivity hotspots’, that are non-conserved binding site residues that control selectivity via favorable target interactions or repulsive ‘off-target’ contacts. It also reviews functional rationale from observed ligand-binding affinities and mutagenesis effects.

Smith KE, Fritzell S, Nilsson A, Barchan K, Rosén A, Schultz L, Varas L, Säll A, Rose N, Håkansson M, von Schantz L, & Ellmark P (2023).
ATOR-1017 (evunzekibart), an Fc-gamma receptor conditional 4-1BB agonist designed for optimal safety and efficacy, activates exhausted T cells in combination with anti-PD-1.
Cancer Immunol. Immunother. https://doi.org/10.1007/s00262-023-03548-7
PDB entry:
8OZ3 – Crystal structure of scFv ATOR 1017 bound to human 4-1BB.
ATOR-1017 binds to a unique epitope on 4-1BB enabling ATOR-1017 to activate T cells, including cells with an exhausted phenotype, and NK cells, in a cross-linking dependent, FcγR-conditional, manner. This translated into a tumor-directed and potent anti-tumor therapeutic effect in vivo, which was further enhanced with anti-PD-1 treatment.

Petruk G, Puthia P, Samsudin F, Petrlova J, Olm F, Mittendorfer M, Hyllén S, Edström D, Strömdahl A-C, Diehl C, Ekström S, Walse B, Kjellström S, Bond PJ, Lindstedt S & Schmidtchen A (2023).
Targeting Toll-like receptor-driven systemic inflammation by engineering an innate structural fold into drugs.
Nat Commun, 14, 6097. https://doi.org/10.1038/s41467-023-41702-y
PDB entries:
8BWW – Targeting Toll-like receptor-driven systemic inflammation by engineering an innate structural fold into drugs
Thrombin-derived C-terminal peptides (TCPs) are endogenous anti-infective immunomodulators interfering with CD14-mediated TLR-dependent immune responses. Using a combination of structure- and in silico-based design, nuclear magnetic resonance (NMR) spectroscopy, biophysics, mass spectrometry, cellular, and in vivo studies, the paper presents the structure, CD14 interactions, protease stability, transcriptome profiling, and therapeutic efficacy of sHVF18.

Billiald P, Slater A, Welin M, Clark JC, Loyau S, Pugnière M, Jiacomini IG, Rose N, Lebozec K, Toledano E, François D, Watson SP & Jandrot-Perrus M (2023).
Targeting platelet GPVI with glenzocimab: a novel mechanism for inhibition
Blood Adv 7, 1258–1268. https://doi.org/10.1182/bloodadvances.2022007863
PDB entries:
7R58 – Crystal structure of the GPVI-glenzocimab complex
Platelet glycoprotein VI (GPVI) is attracting interest as a potential target for the development of new antiplatelet molecules with a low bleeding risk. A cocrystal of glenzocimab with an extracellular domain of monomeric GPVI was obtained and its structure determined to a resolution of 1.9 Å. The data revealed that (1) glenzocimab binds to the D2 domain of GPVI, GPVI dimerization was not observed in the crystal structure because glenzocimab prevented D2 homotypic interactions and the formation of dimers that have a high affinity for collagen and fibrin; and (2) the light variable domain of the GPVI-bound Fab causes steric hindrance that is predicted to prevent the collagen-related peptide (CRP)/collagen fibers from extending out of their binding site and preclude GPVI clustering and downstream signaling.

Nelson, MH, Fritzell S, Miller R, Werchau D, Van Citters D, Nilsson A, Misher L, Ljung Lill, Bader R, Deronic A, Chunyk AG, Schultz L, Varas LA, Rose N, Håkansson M, Gross Jane, Furebring C, Pavlik P, Sundstedt A, Veitonmäki N, Ramos HJ, Säll A, Dahlman A, Bienvenue D, von Schantz L, McMahan CJ, Askmyr M, Hernandez-Hoyos G, Ellmark P (2023).
The Bispecific Tumor Antigen-Conditional 4–1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies
Mol Cancer Ther 22, 89–101. https://doi.org/10.1158/1535-7163.MCT-22-0395
PDB entries:
7YXU – Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB
4–1BB (CD137) is an activation-induced costimulatory receptor that regulates immune responses of activated CD8 T and natural killer cells by enhancing proliferation, survival, cytolytic activity, and IFNγ production. To minimize systemic immune toxicities and enhance activity at the tumor site, the authors have developed a novel bispecific antibody that stimulates 4–1BB function when co-engaged with the tumor-associated antigen 5T4. ALG.APV-527 was built on the basis of the ADAPTIR bispecific platform with optimized binding domains to 4–1BB and 5T4 originating from the ALLIGATOR-GOLD human single-chain variable fragment library. Using X-ray crystallography The epitope of ALG.APV-527 was determined to be located at domain 1 and 2 on 4–1BB.

Zhang Y, Towers CG, Liu J, Håkansson M, Logan DT, Donini O & Kutateladze TG (2022).
Dusquetide modulates innate immune response through binding to p62.
Structure 30, 1055-1061e7. doi: 10.1016/j.str.2022.05.003
PDB entries:
7R1O – p62-ZZ domain of the human sequestosome in complex with dusquetide
SQSTM1/p62 is an autophagic receptor that plays a major role in mediating stress and innate immune responses. Preclinical studies identified p62 as a target of the prototype innate defense regulator (IDR); however, the molecular mechanism of this process remains unclear. The paper describes the structural basis and biological consequences of the interaction of p62 with the next generation of IDRs, dusquetide.
Don’t miss our blog post telling the exciting story behind this publication!

Maurer MF, Lewis JL, Ardour D, Gudgeon CJ, Chandrasekaran S, Mudri S, Kleist, KN, Navas C, Wolfson MF, Rixon MW, Swanson R, Dillon SR, Levin SD, Kimbung YR, Akutsu M, Logan DT, Walse B, Swiderek K & Peng SL (2022).
The engineered CD80 variant fusion therapeutic davoceticept combines checkpoint antagonism with conditional CD28 co-stimulation for anti-tumor immunity.
Nature Commun. 13, 1790. doi:10.1038/s41467-022-29286-5
PDB entries:
7TPS – Crystal structure of ALPN-202 (engineered CD80 vIgD) in complex with PD-L1
The CD80 vIgD from ALPN-202 was co-crystallized with WT PD-L1 ECD and used to determine the structure of the CD80 vIgD:PD-L1 complex. The paper shows that a therapeutic ALPN-202 enhances T cell activation and anti-tumor efficacy in cell-based assays and mouse tumor models more potently than checkpoint blockade alone and thus has the potential to generate potent, clinically meaningful anti-tumor immunity in humans.

Skladanowska K, Bloch Y, Strand J, White K, Hua J, Aldridge D, Welin M, Logan DT, Soete A, Merceron R, Murphy C, Provost M, Bazan JF, Hunter C, Hill J & Savvides SN (2022).
Structural basis of activation and antagonism of receptor signaling mediated by interleukin-27.
Cell Reports, 41, 111490. doi:https://doi.org/10.1016/j.celrep.2022.111490
PDB entries:
7ZXK – Human IL-27 in complex with neutralizing SRF388 FAb fragment
7ZG0 – Murine IL-27 in complex with IL-27Ra and a non-competing Nb
Interleukin-27 (IL-27) uniquely assembles p28 and EBI3 subunits to a heterodimeric cytokine that signals via IL-27Rα and gp130. To provide the structural framework for receptor activation by IL-27 and its emerging therapeutic targeting, the crystal structures of mouse IL-27 in complex with IL-27Rα and human IL-27 in complex with SRF388, a monoclonal antibody undergoing clinical trials with oncology indications, were determined.

Huang S, Xu P, Shen DD, Simon IA, Mao C, Tan Y, Zhang H, Harpsøe K, Li H, Zhang Y, You C, Yu X, Jiang Y, Zhang Y, Gloriam DE, Xu HE (2022).
GPCRs steer Gi and Gs selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol Cell. 82(14):2681-2695.e6. doi:10.1016/j.molcel.2022.05.031
PDB entries:
7XT8 – Serotonin 4 (5-HT4) receptor-Gs-Nb35 complex
7XT9 – Serotonin 4 (5-HT4) receptor-Gs complex
7XTA – Serotonin 4 (5-HT4) receptor-Gi-scFv16 complex
7XTB – Serotonin 6 (5-HT6) receptor-Gs-Nb35 complex
7XTC – Serotonin 7 (5-HT7) receptor-Gs-Nb35 complex
Serotonin (or 5-hydroxytryptamine, 5-HT) is an important neurotransmitter that activates 12 different G protein-coupled receptors (GPCRs) through selective coupling of Gs, Gi, or Gq proteins. The structural basis for G protein subtype selectivity by these GPCRs remains elusive. The paper reports the structures of the serotonin receptors 5-HT4, 5-HT6, and 5-HT7 with Gs, and 5-HT4 with Gi1. The structures reveal that transmembrane helices TM5 and TM6 alternate lengths as a macro-switch to determine receptor’s selectivity for Gs and Gi, respectively.

Ahlqvist J, Linares-Pastén JA, Håkansson M, Jasilonis A, Kwiatkow K, Fridjonsson, OH, Kaczarowska, A-K, Dabrowski, S, Aevarsson, A, Hreggvidsson, GO, Al-Karadaghi S, Kaczorowska, T, Nordberg Karlsson E. (2022).
Crystal structure and initial characterization of a novel archeal-like Holiday junction-resolving enzyme from Thermus thermophilus phage Tth 15-6.
Acta Cryst. D78, 1384-1398. https://doi.org/10.1107/S2059798322009895
Dandare SU, Håkansson M, Svensson LA, Timson DJ, Allen CRC (2022).
Expression, purification and crystallization of a novel metagenome-derived salicylaldehyde dehydrogenase from Apline soil.
Acta Crystallographic Secition F: Structural Biology Communications 78 (4). https://doi.org/10.1107/S2053230X22002345
Kirk, NS, Chen Q, Wu YG, Asante, AL, Hu H, Espinosa JF, Martínez-Olid F, Margetts MB, Faiz A. Mohammed FA, Kiselyov VV, Barrett DG & Lawrence MC (2022)
Activation of the human insulin receptor by non-insulin-related peptides.
Nat Commun 13, 5695. https://doi.org/10.1038/s41467-022-33315-8
The authors acknowledge Carl Diehl for his contribution to the NMR part of the work.
Xu P, Huang S, Zhang H, Mao C, Zhou X E, Cheng X, Simon I A, Shen D-D, Yen H-Y, Robinson C V, Harpsøe K, Svensson B, Guo J, Jiang H, Gloriam D E, Melcher K, Jiang Y, Zhang Y, Xu H E (2021).
Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature 592, 469–473, https://doi.org/10.1038/s41586-021-03376-8
PDB entries: 7E2X, 7E2Y, 7E2Z, 7E32, 7E33
Serotonin, or 5-hydroxytryptamine (5-HT), is a neurotransmitter that activates the largest subtype family of G-protein-coupled receptors (GPCR). Drugs that target 5-HT1A, 5-HT1D, 5-HT1E and other 5-HT receptors are used to treat numerous disorders. The paper reports five structures of 5-HT receptor–G-protein complexes: 5-HT1A in the apo state, bound to 5-HT or bound to the antipsychotic drug aripiprazole; 5-HT1D bound to 5-HT; and 5-HT1E in complex with a 5-HT1E- and 5-HT1F-selective agonist, BRL-54443.

Mujtaba H, Floriane B, Guzelj S, Sundin AP, Håkansson M, Kovačič R, Leffler H, Tomašič T, Anderluh M, Jakopin Ž, Nilsson UJ (2021).
Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain. ACS Med Chem Lett 12(11):1745-1752. doi: 10.1021/acsmedchemlett.1c00371
PDB entries: 7P11, 7P1M
Galectin-8 is a carbohydrate-binding protein that plays a crucial role in tumor progression and metastasis, antibacterial autophagy, modulation of the immune system, and bone remodeling. X-ray structural analysis of galectin-8N in complex with one benzimidazole- and one quinoline-galactal derivative at 1.52 and 2.1 Å together with molecular dynamics simulations and quantum mechanical calculations of galectin-8N in complex with the benzimidazole derivative is presented in this work.

Mujtaba H, van Klaveren S, Håkansson M, Diehl C, Kovačič R, Baussière F, Sundin A P, Dernovšek J, Walse B, Zetterberg F, Leffler H, Anderluh M, Tomašič T, Jakopin Ž, Nilsson U J (2021).
Benzimidazole–galactosides bind selectively to the Galectin-8 N-Terminal domain: Structure-based design and optimisation.
Eu. J. Med. Chem. 223, 113664. https://doi.org/10.1016/j.ejmech.2021.113664
Beckmann R, Jensen K, Fenn S, Speck J, Krause K, Meier A, Röth M, Fauser S, Kimbung R, Logan DT, Steegmaier M & Kettenberger H. (2021).
DutaFabs: A novel platform of bispecific therapeutic Fab fragments which can simultaneously bind two targets with high affinity.
Nature Communications 12, 708. https://doi.org/10.1038/s41467-021-20949-3
Stenström O, Diehl C, Modig K, Nilsson U J, and Akke M. (2021).
Mapping the energy landscape of protein–ligand binding via linear free energy relationships determined by protein NMR relaxation dispersion.
RSC Chem. Biol. 2, 259-265. https://doi.org/10.1039/D0CB00229A
Durcik M, et al. and Welin M, Kimbung R, Focht D et al. and Tomašiča T. (2021).
New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens.
Eur J Med Chem 213, 113200. https://doi.org/10.1016/j.ejmech.2021.113200
Goebel EJ, Kattamuri C, Gipson GR, Krishnan L, Chavez M, Czepnik M, Maguire MC, Grenha R, Håkansson M, Logan DT, Grinberg AV, Sako D, Castonguay R, Kumar R, Thompson TB (2021).
Structures of activin ligand traps using natural sets of type I and type II TGFβ receptors.
iScience 25, 103590. doi: 10.1016/j.isci.2021.103590
Nyerges A, et al. and Welin M, Kimbung R, Focht D, Peterlin Mašič L, and Pal C. (2020). Rational design of balanced dual-targeting antibiotics with limited resistance.
PLoS Biol 18(10):e3000819. https://doi.org/10.1371/journal.pbio.3000819
Karczewski J, Krasucki S P, Asare-Okai P N, Diehl C, Friedman A, Brown C M, Maezato Y, and Streatfield S J. (2020). Isolation, Characterization and Structure Elucidation of a Novel Lantibiotic From Paenibacillus sp.
Front. Microbiol. 24. https://doi.org/10.3389/fmicb.2020.598789
Sanchez-Fernandez A, Diehl C, Houston J E, Leung A E, Tellam J P, Rogers S E, Prevost S, Ulvenlund S, Sjögren H, and Wahlgren M. (2020). An integrative toolbox to unlock the structure and dynamics of protein–surfactant complexes.
Nanoscale Adv 2, 4011-4023. https://doi.org/10.1039/D0NA00194E
Schneider, P., Welin, M., Svensson, B., Walse, B. and Schneider, G. (2020). Virtual screening and design with machine intelligence applied to Pim‐1 kinase inhibitors.
Mol. Inf., 39, 2000109. https://doi.org/10.1002/minf.202000109
Telzerow A, Paris J, Håkansson M, González‐Sabín J, Ríos‐Lombardía N, Gröger H, Morís F, Schürmann M, Schwab H, Steiner K. (2020). Expanding the Toolbox of R‐Selective Amine Transaminases by Identification and Characterization of New Members.
ChemBioChem 2020 21, 1–12. https://doi.org/10.1002/cbic.202000692
Ladds M J G W, Popova G, Pastor-Fernández A, Kannan S, van Leeuwen I M M, Håkansson M, Walse B, Tholander F, Ravi Bhatia, Verma C S, Lane D P, Laín S. (2020). Exploitation of dihydroorotate dehydrogenase (DHODH) and p53 activation as therapeutic targets: A case study in polypharmacology.
J Biol Chem 295, 17935-17949. https://doi.org/10.1074/jbc.RA119.012056
Shilova A, Lebrette H, Aurelius, O, Nan J, Welin M, Kovacic R, Ghosh S, Safari C, Friel R J, Milas M, Matej Z, Högbom M, Brändén G, Kloos M, Shoeman R L, Doak B, Ursby, T M. Håkansson T M, Logan D T, and Mueller U. (2020). Current status and future opportunities for serial crystallography at MAX IV Laboratory
J. Synchrotron Rad. 27, 1095-1102. https://doi.org/10.1107/S1600577520008735
Plotka M, Szadkowska M, Håkansson M, Kovačič R, Al-Karadaghi S, Walse B, Werbowy O, Kaczorowska A-K, and Kaczorowski T. (2020). Molecular Characterization of a Novel Lytic Enzyme LysC from Clostridium intestinale URNW and Its Antibacterial Activity Mediated by Positively Charged N-Terminal Extension.
Int J Mol Sci. 21, 4894; https://doi.org/10.3390/ijms21144894
Baggio C, Udompholkul P, Gambini L, Jossart J, Salem AF, Håkansson M, Perry JJP, Pellecchia M. (2020). N-locking stabilization of covalent helical peptides: Application to Bfl-1 antagonists.
Chem Biol Drug Des. 95, 412-426. doi: 10.1111/cbdd.13661
Telzerow A, Paris J, Håkansson M, Gonzalez-Sabin J, Rios-Lombardía N, Schürmann M, Gröger H, Morís F, Kourist R, Schwab H and Steiner K (2019). Amine Transaminase from Exophiala Xenobiotica – Crystal Structure and Engineering of a Fold IV Transaminase that Naturally Converts Biaryl Ketones.
ACS Catal. 9, 1140−1148. https://doi.org/10.1021/acscatal.8b04524
Dahlqvist A, Mandal S, Peterson K, Håkansson M, Logan DT, Zetterberg FR, Leffler H, Nilsson UJ (2019). 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules. 24 (24), 4554. https://doi.org/10.3390/molecules24244554
Ruggieri F, Campillo-Brocal JC, Chen S, Humble MS, Walse B, Logan DT, Berglund P (2019). Insight into the dimer dissociation process of the Chromobacterium violaceum (S)-selective amine transaminase.
Sci Rep. 9, 16946. DOI: 10.1093/nar/25.17.3389
Freitag-Pohl S, Jasilionis A, Håkansson M, Svensson LA, Kovačič R, Welin M, Watzlawick H, Wang L, Altenbuchner J, Płotka M, Kaczorowska AK, Kaczorowski T, Nordberg Karlsson E, Al-Karadaghi S, Walse B, Aevarsson A, Pohl E (2019). Crystal structures of the Bacillus subtilis prophage lytic cassette proteins XepA and YomS.
Acta Crystallogr D Struct Biol., 75(Pt 11):1028-1039. https://doi.org/10.1107/S2059798319013330
Korkmaz B, Lesner A, Wysocka M, Gieldon A, Håkansson M, Gauthier F, Logan DT, Jenne DE, Lauritzen C, Pedersen J (2019). Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem Pharmacol. 164, 349-367. https://doi.org/10.1016/j.bcp.2019.04.006
Kracht ON, Correia Cordeiro RS, Håkansson M, Stockmann J, Sander D, Bandow J, Senges CHR, Logan DT, Kourist R (2019). Discovery of three novel sesquiterpene synthases from Streptomyces chartreusis NRRL 3882 and crystal structure of an α-eudesmol synthase.
J Biotechnol. 297, 71-77. https://doi.org/10.1016/j.jbiotec.2019.03.006
Casaletto JB, Geddie ML, Abu-Yousif AO, Masson K, Fulgham A, Boudot A, Maiwald T, Kearns JD, Kohli N, Su S, Razlog M, Raue A, Kalra A, Håkansson M, Logan DT, Welin M, Chattopadhyay S, Harms BD, Nielsen UB, Schoeberl B, Lugovskoy AA, MacBeath G (2019). MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc Natl Acad Sci U S A 116, 7533-7542. https://doi.org/10.1073/pnas.1819085116
Ruggieri F, van Langen LM, Logan DT, Walse B, Berglund P. (2018). Transaminase-Catalyzed Racemization with Potential for Dynamic Kinetic Resolutions.
ChemCatChem, 10, 1-8. https://doi.org/10.1002/cctc.201801049
Gustafsson NMS, Färnegårdh K, Bonagas N, Ninou AH, Groth P, Wiita E, Jönsson M, Hallberg K, Lehto J, Pennisi R, Martinsson J, Norström C, Hollers J, Schultz J, Andersson M, Markova N, Marttila P, Kim B, Norin M, Olin T, Helleday T (2018). Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination.
Nat Commun. 9, 3872. DOI: 10.1038/s41467-018-06287-x
Abdillahi SM, Maaß T, Kasetty G, Strömstedt AA, Baumgarten M, Tati R, Nordin SL, Walse B, Wagener R, Schmidtchen A, Mörgelin M (2018)
Collagen VI Contains Multiple Host Defense Peptides with Potent In Vivo Activity.
J Immunol., 201, 1007-1020. https://doi.org/10.4049/jimmunol.1700602
Ladds MJGW, van Leeuwen IMM, Drummond CJ, Chu S, Healy AR, Popova G, Pastor Fernández A, Mollick T, Darekar S, Sedimbi SK, Nekulova M, Sachweh MCC, Campbell J, Higgins M, Tuck C, Popa M, Safont MM, Gelebart P, Fandalyuk Z, Thompson AM, Svensson R, Gustavsson AL, Johansson L, Färnegårdh K, Yngve U, Saleh A, Haraldsson M, D’Hollander ACA, Franco M, Zhao Y, Håkansson M, Walse B, Larsson K, Peat EM, Pelechano V, Lunec J, Vojtesek B, Carmena M, Earnshaw WC, McCarthy AR, Westwood NJ, Arsenian-Henriksson M, Lane DP, Bhatia R, McCormack E, Laín S. A (2018). A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage.
Nat Commun. 9, 1107. DOI: 10.1038/s41467-018-03441-3
Andersen MCF, Boos I, Kinnaert C, Awan SI, Pedersen HL, Kračun SK, Lanz G,Rydahl MG, Kjærulff L, Håkansson M, Kimbung R, Logan DT, Gotfredsen CH, Willats WGT, Clausen MH (2018). Synthesis of branched and linear 1,4-linked galactan oligosaccharides.
Org Biomol Chem. 16, 1157-1162. DOI: 10.1039/c7ob03035e
Peterson K, Kumar R, Stenström O, Verma P, Verma PR, Håkansson M, Kahl-Knutsson B, Zetterberg F, Leffler H, Akke M, Logan DT, Nilsson UJ (2018). Systematic tuning of fluoro-galectin-3 interactions provides thiodigalactoside derivatives with single digit nM affinity and high selectivity.
J Med Chem 61, 1164–1175. DOI: 10.1021/acs.jmedchem.7b01626.
Zetterberg FR, Peterson K, Johnsson R, Brimert T, Håkansson M, Logan DT, Leffler H, Nilsson UJ (2018). Monosaccharide derivatives with low nM lectin affinity and high selectivity based on combined fluorine-amide, phenyl-arginine, sulfur-π, and halogen bond interactions.
ChemMedChem 13, 133-137. DOI: 10.1002/cmdc.201700744.
Takemoto Y, Slough DP, Meinke G, Katnik C, Graziano ZA, Chidipi B, Reiser M, Alhadidy MM, Ramirez R, Salvador-Montañés O, Ennis S, Guerrero-Serna G, Haburcak M, Diehl C, Cuevas J, Jalife J, Bohm A, Lin YS, Noujaim SF (2017). Structural basis for the antiarrhythmic blockade of a potassium channel with a small molecule.
FASEB J pii: fj.201700349R. DOI: 10.1096/fj.201700349R.
Anderson LC, Håkansson M, Walse B and Nilsson CL (2017). Intact Protein Analysis at 21 Tesla and X-Ray Crystallography Define Structural Differences in Single Amino Acid Variants of Human Mitochondrial Branched-Chain Amino Acid Aminotransferase 2 (BCAT2).
J Am Soc Mass Spectrom 28, 1796-1804. DOI: 10.1007/s13361-017-1705-0
Walse B, Turnbull AP and Boyd SM (2017). Tailoring Hit Identification and Qualification Methods for Targeting Protein–Protein Interactions.
In Applied Biophysics for Drug Discovery (Huddler, D. and Zartler, E.R., ed.) pp. 29-59, John Wiley & Sons, Inc, Hoboken, NJ, USA.
Noresson AL, Aurelius O, Öberg CT, Engström O, Sundin AP, Håkansson M, Stenström O, Akke M, Logan DT, Leffler H, Nilsson UJ (2017).
Designing interactions by control of protein-ligand complex conformation: tuning arginine-arene interaction geometry for enhanced electrostatic protein-ligand interactions.
Chem Sci. 9, 1014-1021.
Nilsson LM, Green LC, Veppil Muralidharan S, Demir D, Welin M, Bhadury J, Logan D, Walse B and Nilsson JA (2016). Cancer differentiating agent hexamethylene bisacetamide inhibits BET bromodomain proteins.
Cancer Res. 76, 2376-2383. DOI: 10.1158/0008-5472.CAN-15-2721
Badarau A, Rouha H, Malafa S, Battles MB, Walker L, Nielson N, Dolezilkova I, Teubenbacher A, Banerjee S, Maierhofer B, Weber S, Stulik L, Logan DT, Welin M, Mirkina I, Pleban C, Zauner G, Gross K, Jägerhofer M, Magyarics Z & Nagy E (2016). Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies.
MAbs. 8, 1347-1360. DOI: 10.1080/19420862.2016.1215791
Badarau A, Rouha H, Malafa S, Logan DT, Håkansson M, Stulik L, Dolezilkova I, Teubenbacher A, Gross K, Maierhofer B, Weber S, Jägerhofer M, Hoffmann D, Nagy E (2015). Structure-Function Analysis of Heterodimer Formation, Oligomerization, and Receptor Binding of the Staphylococcus aureus Bi-component Toxin LukGH.
J. Biol. Chem. 290, 142-156. DOI: 10.1074/jbc.M114.598110
Rodrigues T, Reker, D, Welin M, Caldera M, Brunner C, Gabernet G,Schneider P, Walse B and Schneider G (2015). De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angewandte Chemie International Edition, DOI: 10.1002/anie.201508055.
Fisher SZ, von Schantz L, Håkansson M, Logan DT & Ohlin M. (2015). Neutron Crystallographic Studies Reveal Hydrogen Bond and Water-Mediated Interactions between a Carbohydrate-Binding Module and Its Bound Carbohydrate Ligand.
Biochemistry 54, 6435-6438. DOI: 10.1021/acs.biochem.5b01058
Jong-Eun Kim, Joe Eun Son, Hyein Jeong, Dong Joon Kim, Sang Gwon Seo, Eunjung Lee, Tae Gyu Lim, Jong Rhan Kim, Yengo Raymond Kimbung, Hanyong Chen, Ann M. Bode, Ki Won Lee and Zigang Dong (2015). A Novel Cinnamon-Related Natural Product with Pim-1 Inhibitory Activity Inhibits Leukemia and Skin Cancer.
Cancer Res. 75, 2716-2728. doi: 10.1158/0008-5472.CAN-14-3655
von Schantz L, Håkansson M, Logan DT, Nordberg-Karlsson E and Ohlin M (2014). Carbohydrate binding module recognition of xyloglucan defined by polar contacts with branching xyloses and CH-π interactions.
Proteins, 82, 3466-75.
Turnbull AP, Boyd SM & Walse B (2014). Fragment-based drug discovery and protein-protein interactions.
Res. Rep Biochem. 4, 13-26.
Lolli ML, Ducime A, Federico, A Pippione AC, Sainas S, Barge A, Martina K, Boschi D, Lupino E, Piccinini M, Kubbutat M, Schächtele C, Contreras JM, Morice C, Sussman J, Peleg J, Walse B and Al-Kadaraghi S (2014). Towards a Bioisosteric Alkaest: application to the bioisosteric modulation of IMD-0354.
In: Book of Abstracts. p. 234, Lisbon, Portugal, September 7-11. See poster…
Saraboji K, Håkansson M, Genheden S, Diehl C, Qvist J, Weininger U, Nilsson UJ, Leffler H, Ryde U, Akke M & Logan DT. (2012). The Carbohydrate-Binding Site in Galectin-3 Is Preorganized To Recognize a Sugarlike Framework of Oxygens: Ultra-High-Resolution Structures and Water Dynamics.
Biochemistry 2012, 51:296-306.
Humble MS, Cassimjee KE, Håkansson, M, Kimbung YR, Walse B, Abedi V, Federsel H-J, Berglund P & Logan DT (2012). Crystal structures of the Chromobacterium violaceum ω-transaminase reveal major structural rearrangements upon binding of coenzyme PLP.
FEBS J., 279, 779-792.
von Schantz, L., Håkansson, M., Logan, D.T., Walse, B., Osterlin, J., Nordberg-Karlsson, E. and Ohlin, M. (2012). Structural basis for carbohydrate-binding specificity -a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 948-961.
Boyd SM, Turnbull AP and Walse B (2012). Fragment library design considerations.
WIREs Comput. Mol. Sci., 2, 868-885.
Kasetty G, Papareddy P, Kalle M, Rydengård V, Walse B, Svensson B, Mörgelin M, Malmsten M & Schmidtchen A (2011). The C-terminal sequence of several human serine proteases encodes host defense functions.
J Innate Immun. 3, 471-482.
Svensson SL, Pasupuleti M, Walse B, Malmsten M, Mörgelin M, Sjögren C, Olin AI, Collin M, Schmidtchen A, Palmer R & Egesten A (2010). Midkine and pleiotrophin have bactericidal properties: preserved antibacterial activity in a family of heparin-binding growth factors during evolution.
J Biol Chem. 285, 16105-16115.
Diehl C, Engström O, Delaine T, Håkansson M, Genheden S, Modig K, Leffler H, Ryde U, Nilsson UJ & Akke M (2010). Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3.
J Am Chem Soc. 132, 14577-1489.
Fritzson I, Svensson B, Al-Karadaghi S, Walse B, Wellmar U, Nilsson UJ, da Graça Thrige D & Jönsson S. (2010).
Inhibition of human DHODH by 4-hydroxycoumarins, fenamic acids, and N-(alkylcarbonyl)anthranilic acids identified by structure-guided fragment selection.
ChemMedChem. 5, 608-617.
