Protein Crystallization & Protein Crystallography Services: Practical Aspects
With our extensive expertise in protein crystallization and crystallography services and experience working with many different protein classes, we excel in delivering high-quality results for various structural biology and drug discovery projects.
Crystallization & Crystallography workflow
Below, we outline the typical workflow for our protein crystallization and protein crystallography services. Our project types include gene-to-structure, protein-ligand co-crystallization, antibody, and antibody-antigen complex crystallization and structure determination. Leveraging our libraries of off-the-shelf structures (FastLane™ Premium and FastLane™ Standard), we accelerate our client’s projects by offering hundreds of protein targets with validated expression, purification, and crystallization protocols. Sometimes, clients prefer to send samples produced in their laboratories. The information on this page and our sample handling and shipping guide can provide helpful details on preparing and sending samples
If you are ready to benefit from our protein X-ray crystallography services, please don’t hesitate to contact us via the provided contact form for further details.
Protein Crystallization – Sample Preparation
Several essential requirements must be fulfilled to obtain crystallization-grade protein to ensure the protein crystallography project is successful. These requirements are general, we provide them here in case a client decides to send own sample for crystallization and structure determination.
First, the protein must be well-folded, stable, and soluble within a given temperature, concentration, and pH range. Before crystallization, the solution must be monodisperse, i.e., containing no denatured or aggregated material. The protein solution may be characterized using the following biophysical methods:
- Dynamic Light Scattering (DLS)
- Circular dichroism (CD) spectroscopy
- Differential Scanning Fluorimetry (DSF, similar to thermal shift assay)
- Protein NMR spectroscopy
Monodispersity, which DLS assesses, is crucial in protein crystallization. CD spectroscopy is used to verify secondary structure content to ensure that the structure of the purified protein is intact. In contrast, DSF is used to assess the molecule’s stability in different buffer solutions and at different temperatures. This helps in the preparation of the solutions for crystallization & crystallography. In NMR spectroscopy, the HSQC fingerprint spectrum shows whether the protein is well-folded, unfolded at a given temperature, or if some parts of the molecule are highly flexible. Our staff follows specific protocols during all the experiments to maximize their efficiency. The results of the measurements are subsequently used to decide whether the protein is suitable for crystallization and crystallography or if some modifications in the cloning and purification protocols are necessary.
An example of a product sheet for one of our crystallization-grade Protein Shop proteins can be downloaded here.

Identifying the Crystallization Conditions
We can start protein crystallization if we are satisfied with the protein’s state in the solution. However, many other factors may potentially affect crystallization. For this reason, in a crystallography project, we must run screens to identify the best conditions for obtaining well-diffracting crystals.
Our protein crystallization and crystallography services workflow starts by running a large number of screens. Commercial screens, like those from Hampton Research or Molecular Dimensions, are initially used. In addition, the initial conditions usually need to be optimized to obtain well-diffracting single crystals for subsequent structure determination. The large number of buffer conditions used in the screening reflects the large number of parameters and combinations of the parameters that may affect the crystallization. The most common protocols rely on the variation of the different conditions, the most common being:
- Type of buffer and its pH
- Ionic strength
- The presence of various salts in the solution
- The presence of ligands (co-factors, substrate analogs, inhibitors)
- The type of precipitant used (polyethylene glycol (PEG) and ammonium sulfate are the most common), etc.
Our protein crystallization and crystallography services routinely use high-throughput and high-precision liquid handling and imaging robotics to set up the initial screens and optimize newly identified conditions. With robotics, we can screen 96 conditions using as little as 15 microL of protein sample, saving time and precious material. The crystallization plates with the drops are stored in plate hotels at a constant temperature to avoid temperature fluctuations, and they are monitored using a computerized optical system. If you need more information on our services, please get in touch with us using the contact form.


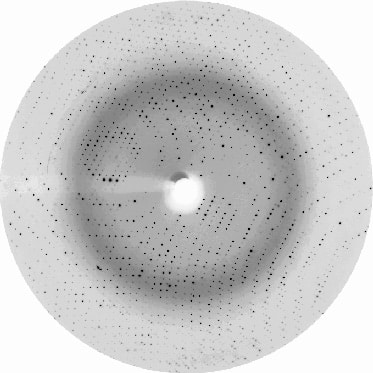
The Crystalography After Crystallization: X-ray Diffraction Data Collection And Structure Determination
Protein crystallization is only the first step in a crystallography project. Once we have obtained the crystals, we will test them in an X-ray beam to collect X-ray diffraction data. For our crystallography services projects we primarily collect X-ray diffraction data at the BioMax beamline at MAX IV Laboratory, one of the world’s best synchrotrons, conveniently located just a few kilometers from our labs in Lund. Our crystallography services also include sending the crystals to other synchrotrons in Europe.
Crystalography needs phase information to determine the protein structure. The phases cannot be obtained directly from the X-ray diffraction pattern. When a protein has been crystallized and an experimental structure already exists, in crystallography the method of molecular replacement is used to obtain the phase information. These phases enable us to calculate an electron density map to build a protein model. We then refine the model to ensure that the finer details of the structure, such as the positions of protein and ligand atoms, are well defined. Our services platform is optimized to carry out all these processes promptly and efficiently.
Molecular replacement significantly accelerates the protein crystallography process. In some instances, models generated by the AlphaFold project can also be utilized for initial phasing. However, in cases where a reliable model is not available, experimental methods are employed to obtain phase information. Typically, a selenomethionine (SeMet)-labeled protein sample, which can be produced upon client’s request, enables structure determination using synchrotron radiation in conjunction with the method of anomalous scattering of X-rays. This means that a (SeMet-labeled sample will be required for the protein crystallization and crystallography services project.
Please get in touch with us directly or visit our X-ray crystallography services page for more details.
