Antibody Structure & Biosimilars Comparability Analysis Services
SARomics Biostructures’ team possesses extensive expertise in monoclonal antibody structure and comparability analysis of biosimilars, including X-ray structure characterization of antibody-antigen complexes and comparability studies of biosimilars’ higher-order structure (HOS) using 2D-NMR spectroscopy.

Antibody And Antibody-Antigen Complex Structure
We can assist you in determining the 3D structure of antibodies and Fab-antigen complexes. Through our gene-to-structure capabilities, we can also help you produce the antigen.
Antibody Structure & Comparability Analysis
SARomics Biostructures’ team has extensive experience in antibody structure and biosimilars comparability analysis by X-ray crystallography and NMR spectroscopy. The structural characterization and comparability studies of biosimilars’ higher-order structure (HOS) uses 13C-natural abundance 2D-NMR spectroscopy. All our studies include biophysical characterization of the state of the monoclonal antibodies and biosimilars in solution using biophysical methods.
More detailed, monoclonal antibody structure services can help you with the following:
- Epitope definition to file stronger IP.
- Understanding the mode of action.
- Structure-based antibody engineering: affinity maturation, humanization, antibody-drug conjugates (ADC), etc.
- Structural characterization of biologics for regulatory purposes.
- Assessing the comparability of a biosimilar antibody and its reference product.
Explore our list of Extracellular Proteins
For antibody-antigen co-crystallization, please find below a list of our FastLane™ Premium and FastLane™ Standard extracellular proteins targeted by therapeutic antibodies. The proteins from the Premium list are available in our laboratory for immediate co-crystallization, while those from the standard list can be expressed and crystallized within a few weeks. You can also view the complete list of FastLane™ structures.
Featured Structures: Platelet Glycoprotein VI (GPVI)
Several animal experiments have demonstrated that platelet glycoprotein VI (GPVI) contributes to thrombosis in ischemic stroke. On this basis, GPVI is a potential target for developing new antiplatelet molecules with low bleeding risk. The crystal structure of Glenzocimab, a Fab fragment of humanized anti-GPVI monoclonal antibody, with the monomeric extracellular domain of Platelet glycoprotein VI GPVI, has been determined at 1.9 Å. The structure shows that Glenzocimab binds to the site of dimerization in the D2 domain of GPVI and inhibits GPIV interaction with CRP, collagen, and fibrin by loss of dimerization site, conformational changes, and steric hindrance.
Antibody Structure and Comparability Analysis Using NMR Spectroscopy
Assessing antibody structure and comparability analysis is critical to developing new products that adhere to patient safety principles (see our blog post on the subject). The NMR-based approach offered by SARomics Biostructures is currently the most efficient HOS comparability assessment approach at atomic resolution. Please see our blog post on comparing the experimental methods for assessing a biosimilar’s structural comparability.
At SARomics Biostructures, we use advanced NMR spectroscopy, which makes it possible to acquire a unique fingerprint representation of the 3D conformation of large, complex molecules like biologics. For biosimilar comparability assessment, 2D 13C NMR spectra of the methyl region of the biomolecules are acquired, and the biosimilar and originator spectra are compared. The method is based on natural abundance and does not require additional expensive labeling. By matching the NMR fingerprint of a given protein to its high-resolution 3D structure, determined, e.g., by X-ray crystallography, or to a fingerprint of another protein batch of the therapeutic monoclonal antibody or its biosimilar, we can rapidly assess and analyze comparability and show that the molecules, for example, a biosimilar and its originator, or different batches or alternative preparations of the same biologic, have identical HOS. In addition, the biomolecules can be studied in the formulation buffer for formulation optimization and batch comparison. Depending on the biomolecule, around 1-2 mg of protein can be sufficient for HOS analysis.
The presentation below provides details of the method and a case study.
Case Studies: SARomics’ Contribution to Antibody-Antigen Complex Structure Determination
Below is a list of published work involving the crystallization and structure determination of antibody-antigen complexes contributed by SARomics Biostructures. The papers have been published in reputable journals such as PNAS, Nature Communications, Cell Reports, iScience, Cancer Therapy, Blood Advances, and Structure. Some of the recent papers are listed below; more can be found in the pdf on the right.
- Structural analysis of light chain-driven bispecific antibodies targeting CD47 and PD-L1. Malinge (Light Chain Bioscience – Novimmune SA) et al., 2024, mAbs, 16, 2362432.
- Structure-guided engineering of immunotherapies targeting TRBC1 and TRBC2 in T cell malignancies. Ferrari (Autolus Therapeutics) et al., 2024, Nat Commun, 15.
- SRF388 Fab in complex with IL-27. Skladanowska (Surface Oncology Inc.) et al., 2022. Cell Reports, 41, 111490.
- Structure-based engineering of a novel CD3ε-targeting antibody for reduced polyreactivity. Liu (Adimab, LLC) et al., 2023, MAbs, 15, 2189974-21899.
- Structures of activin ligand traps using natural sets of type I and type II TGFβ receptors. Goebel (Acceleron Pharma) et al., 2022. iScience, 25, 103590.
- The bispecific 4-1BB x 5T4 agonist, ALG.APV-527, mediates strong T-cell activation and potent anti-tumor activity. Nelson (Alligator Bioscience AB) et al., 2022, Mol. Cancer Ther., 22-0395.
- Targeting platelet GPVI with glenzocimab: a novel mechanism for inhibition. Billiald (Acticor Biotech) et al., 2022. Blood Adv (2023) 7 (7): 1258–1268.
- DutaFab in complex with its antigens PDGF and VEGFA. Beckmann (Roche) et al., 2021. Nat Comm, 12:708.
- Bispecific Antibody Mediating PD-L1–Dependent CD28 Co-stimulation on T Cells for Enhanced Tumor Control. Majocchi (Light Chain Bioscience – Novimmune SA) et al., 2024. Cancer Immunol Res, 0298.
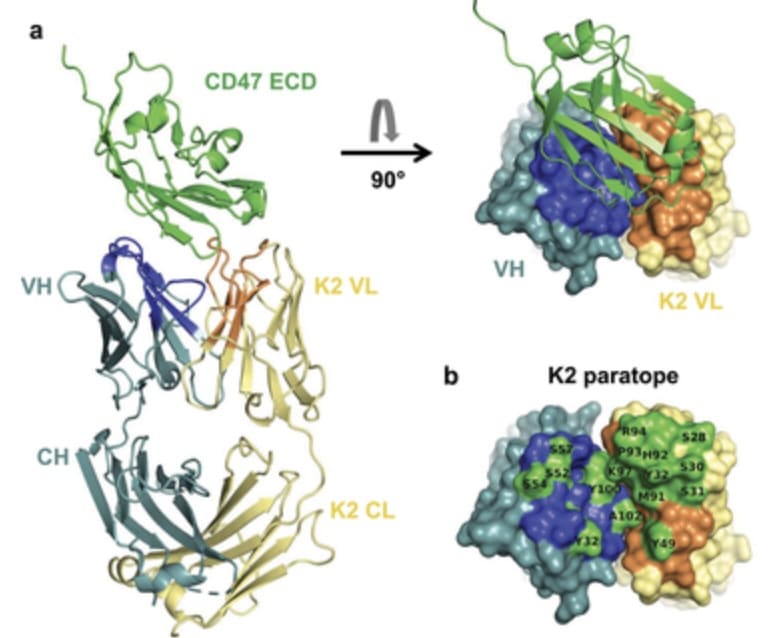
Recent Publication: The structure of unique light chain-driven bispecific antibodies
P Malinge, X Chauchet, J Bourguignon, N Bosson, S Calloud, T Bautzova, M Borlet, M Laursen, V Kelpsas, N Rose, F Gueneau, U Ravn, G Magistrelli & N Fischer (2024).
Structural analysis of light chain-driven bispecific antibodies targeting CD47 and PD-L1. mAbs 16, NO. 1, 2362432.
https://doi.org/10.1080/19420862.2024.2362432
The authors developed a bispecific antibody format, which, unlike natural antibodies that mainly rely on the heavy chain (HC), drives antigen binding and specificity using the light chain (LC). To better understand epitope-paratope interactions in the antibody-antigen complex, the SARomics team determined the X-ray crystallographic structures of an antigen-binding fragment (Fab) in a complex with human CD47 and another Fab in a complex with human PD-L1 (one of the proteins in our FastLane™ Premium library). Structure analysis revealed the dominant contribution of the light chain, demonstrating that the light chain can also mediate high-affinity binding, enabling the creation of bispecific antibodies with native structures, thus enhancing the therapeutic potential. (PDB IDs: 8RP8 and 8RPB).
We invite you to visit our publications page for additional examples of projects we have contributed to.